Single cell-based computational tool for subpopulation conversion
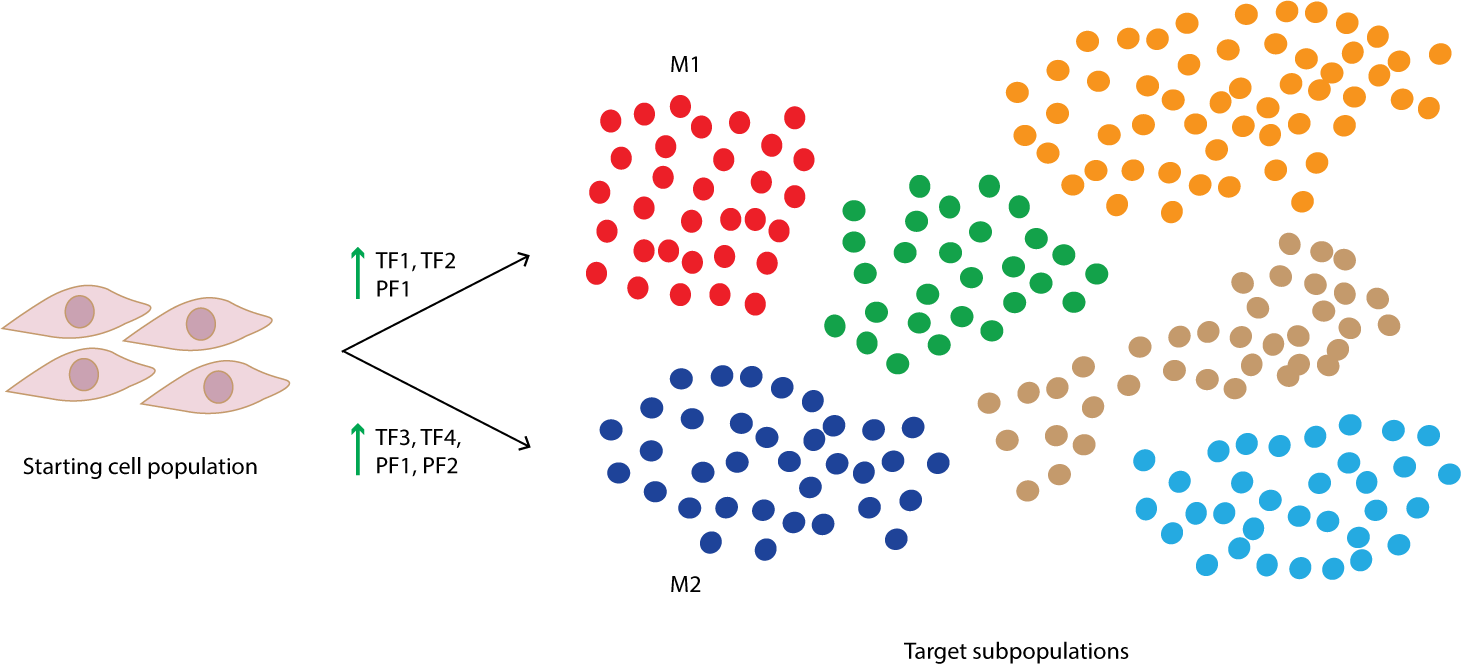
Convert between cell subtypes by using single-cell data to identify an optimal, subtype-specific set of core transcription factors personalized for your own human or mouse cell study.
Examples: Please select "Preview Examples" button in the main page to find gene expression matrix and cluster information format examples, obtained from La Manno et al. 2016 data.
The results are compressed in a .zip file. Inside, you will find a summary file that contains the metadata of your analysis, an hierarchical clustering dendrogram of your gene expression matrix, and two tables: "cores.tsv” containing the predicted cell conversion TFs for each target subpopulation, ranked by the fold-change; and “markers.csv” containing the top 10 predicted marker genes of each target subpopulation, ranked by JSD score (low JSD = unique and specific marker).
This software was developed in the Computational Biology Group by Mariana Ribeiro, Dr. Satoshi Owaka and Prof. Dr. Antonio del Sol.
Within any publication that uses any methods or results derived from or inspired by TransSynW, please cite:
Ribeiro MM, Okawa S, del Sol A. TransSynW: A single-cell RNA-sequencing based web application to guide cell conversion experiments. STEM CELLS Transl Med. 2020;1–9.